Hardware Solutions
Applications
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Development
The research groups of Drs. Bally-Cuif and Emmanuel Beaurepaire, from the Paris-Saclay Institute for Neuroscience and Ecole Polytechnique, have developed a method that allows imaging of adult neural stem cells in their intact physiological context over long periods. The new approach uses Imaris software to achieve dynamic analysis of in vivo neural stem cell behavior.
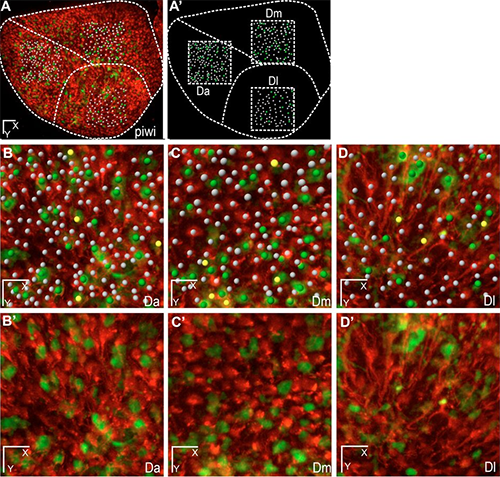
Figure 1. These whole-mount dorsal views show three areas of the neural stem cell layer of one hemisphere of the zebrafish adult pallium (positioned in A,A’ on the stem cell layer) with plotted quiescent neural stem cells (while dots), dividing neural stem cells (yellow dots), and dividing non-stem progenitors (green dots). On the live images, neural stem cells are highlighted in red by the her4:drfp transgene, and dividing cells in green by the mcm5:egfp transgene. Scale bars: 50 μm (C), 15 μm (D). Reprinted from Dray et al. Development. 142: 3592-3600.
Adult brains of vertebrates contain sites where neurons of high physiological impact are generated from adult neural stem cells. Despite the importance of these cells, the mechanisms and pathways controlling the maintenance and recruitment of adult neural stem cells are still incompletely understood. This knowledge gap is largely tied to the challenges involved in analyzing neural stem cell pools in situ. Culturing cells or brain slices in vitro is likely to alter stem cell properties profoundly, thus the researchers set out to develop a way to study these cells in their natural environment.
Long-term observations
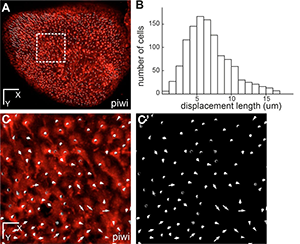
Figure 2. This whole-mount dorsal view of the neural stem cell layer of one hemisphere of the zebrafish adult pallium where all individual neural stem cells have been plotted (white dots, using the her4:drfp transgene as in Fig.1) (fish named piwi). B- Distribution of the total displacement length of each cell after 7 days of imaging. C- Overall displacement vector of each individual cell after 7 days of imaging in the area boxed in A (C: vectors shown on top of red fluorescence –neural stem cells-, C’: vectors only). Scale bars: 80 μm (A), 20 μm (C-C’). Reprinted from Dray et al. Development. 142: 3592-3600.
The researchers developed an experimental workflow that combined multicolor fluorescence with second- and third-harmonic multiphoton microscopy for observing adult neural stem cells in the brain of adult zebrafish over days or weeks. Fluorescence signals were used to detect adult neural stem cell markers, while endogenous second- and third-harmonic generation signals provide morphological landmarks for longitudinal imaging on different days and for data registration.
The investigators used this approach to image adult neural stem cells through the skin and skull of a live anesthetized zebrafish every two to four days and then used Imaris to realign the large 3D images acquired over weeks. Imaris also provided 3D segmentation and tracking with manual correction, which were critical to the new imaging approach.
“The most critical capability of Imaris was the ImarisXT interface, which let us use RecursiveReg, an ImarisXT function provided by Michael Liebling for automated pixel-based registration,” Drs. Dray and Willy Supatto say. “We used it to obtain a coarse alignment of the brain images using the harmonic signals. Image alignment was then refined after tracking a few cells by using the drift correction function of Imaris.” Dr. Supatto adds that ImarisXT was also useful for exporting tracking results and computing 3D image correlations with Matlab.
Characterizing neural stem cells
The researchers’ new approach enabled the identification and tracking of thousands of adult neural stem cells with single cell resolution. They used these images to characterize the in vivo distribution, frequency, and mode of adult neural stem cell division events in their endogenous niche. They found that neural stem cells in distinct areas differ in several parameters, including cell density and activation frequency. “Adult neural stem cells in the zebrafish brain are highly similar to their rodent counterparts, and the results obtained in the zebrafish system, so far, have been fully generalizable,” says Dr. Bally-Cuif.
In addition to studying adult neural stem cells, the new image analysis approach can be used to track adult neural stem cell behavior in other brain areas and under manipulated conditions such as lesions or drug application. It can also be used to track any type of cell that is labeled by a fluorescent transgene in the brain or any tissue located relatively superficially in the body.
Author: Drs. Laure Bally-Cuif and Nicolas Dray, Paris-Saclay Institute for Neuroscience Drs. Emmanuel Beaure
Category: Case Study
