Hardware Solutions
Applications
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Cell Biology
Dr. Yaron Shav-Tal and lab members, Bar-Ilan University in Israel
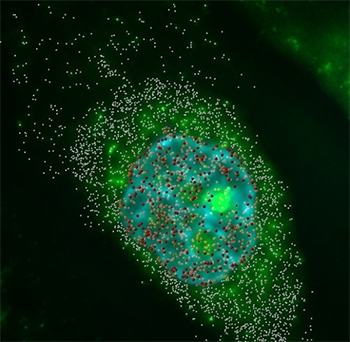
Messenger RNAs were imaged in three dimensions and deconvolved. By distinguishing the location of the mRNA, the researchers could calculate the cytoplasm to nucleus ratio. Red spot objects indicate mRNA particles in the nucleus, and the white spheres show those in the cytoplasm. Hoechst DNA counterstaining is shown in blue. Videos and images acquired by Amir Mor.
During gene expression messenger RNA moves from the nucleus to the cytoplasm and although this process has been studied from a biochemical point of view, very little is known about its kinetics. Imaris software helped researchers from Bar-Ilan University in Israel gain in vivo insight by letting them visualize and quantify the kinetics of mRNA transport in single living cells.
Most measurements of RNA export have been performed in fixed cells or with in vitro assays. To study the in vivo kinetics of mRNA export and to track mRNAs during export, the researchers set up a unique cell system. The cells had stably integrated gene constructs that generated complementary mRNA transcripts that could be observed in cells using a fluorescent protein.
“This system enabled us to perform detailed kinetic analysis of different levels of mRNA export and live-imaging of mRNA translocation from the site of transcription through association with the nuclear pore complex, translocation through the pore and release into the cytoplasm - all at the single mRNA level,” said Amir Mor, the graduate student who led this research.
The researchers used Imaris, the most powerful and versatile 3D and 4D image analysis software solution on the market and its spot object analysis to track single mRNA molecules. The ImarisTrack module automatically identified mRNA spots in each time frame and connected each spot to produce tracks that showed the coordinates of an individual mRNA molecule over time. From the coordinates they could characterize the type of mRNA movement as well as its speed. This was repeated for tracking of several different mRNA species in two dimensions as well as in three dimensions.

The researchers used Imaris to visualize the location of messenger RNAs molecules (spheres – Imaris spot objects) in three dimensions. Videos and images acquired by Amir Mor.
The researchers then combined the 3D rendering of labeled mRNA spots with stained chromatin images to capture the biophysical nature of mRNA movement, which takes place in regions where the chromatin is less condensed. The nuclear staining also allowed them to determine whether an mRNA resided in the nucleus or cytoplasm and thus to further compare their movements. After classifying the location of the mRNA particles, the researchers determined the ratio of mRNAs in the cytoplasm to those in the nucleus and calculated the export efficiencies for the different mRNA species.
The researchers quantified the kinetic contribution of each step in the gene expression pathway, studied mRNA translocation on the single mRNA level, provided the first measurements of mRNA export through the nuclear pore, and dissected the different steps of mRNA anchoring and translocation at the nuclear pore complex in living cells. Importantly, they also proposed the mechanism by which nucleoplasmic mRNAs move with respect to the chromatin.
“Altogether, our work has provided the first extensive outlook on the kinetics of mRNA nucleo-cytoplasmic transport and export, from the time of transcription to cytoplasmic localization,” Dr. Shav-Tal concludes.
Research Paper: Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells, Nature Cell Biology 12, 543 - 552 (2010) doi:10.1038/ncb2056.
