Hardware Solutions
Applications
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Neuroscience
Dr. Gary J. Bassell and lab members, Emory University, US
Dr. Gary J. Bassell and members of his lab from Emory University recently developed an automated method that uses Imaris image processing and analysis software to study dendritic spine morphology. Uncovering the mechanisms regulating dendritic spine morphology is critical for understanding the basic properties of neuronal development and function.
“Manual spine analysis is time consuming and difficult to reproduce,” said Sharon Swanger, a graduate student in the Bassell lab. “We needed an automated program that could detect and measure spines in 3D and be applied to the large studies necessary to discern the mechanisms regulating spine morphology.”
The researchers had successfully performed volumetric analyses with other Imaris modules, so they decided to test whether the Filament Tracer module could accurately trace and measure dendritic spines. “We compared different analysis programs and labeling methods and found that Filament Tracer combined with Lifeact-expressing neurons provided the most consistent and accurate dendritic spine analyses for large datasets,” Swanger said.
With Imaris, the most powerful and versatile 3D and 4D image analysis software solution on the market for researchers in life sciences and its module Filament Tracer, a single set of input parameters can be used to automatically detect and measure spines across a large dataset. Filament Tracer also provides a wealth of statistics for spine number and spine geometry and the ability to track these measures for individual spines across time.
For dendritic spine analysis, the researchers used Imaris’s Surpass view to create a new filament in Autopath mode and select part of a dendrite as a region of interest (ROI). For time-lapse imaging, they assigned a single dendrite starting point at each timepoint using the AutoDepth mode of Filament Tracer. They traced the spines by setting the maximum spine length and minimum spine end diameter and then using automatic thresholds to generate spine seed points and surface rendering. After generating a trace, they applied a filter to ensure all dendritic protrusions were assigned as spines.
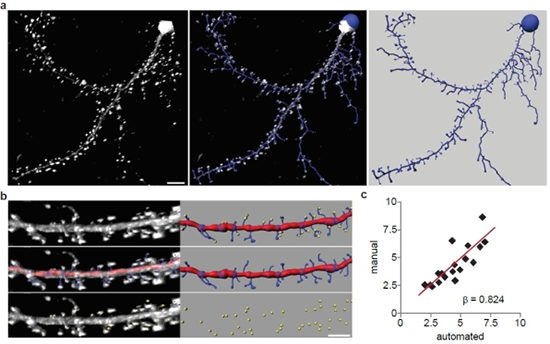
The researchers performed automated detection of dendritic spines using images of Lifeact-expressing neurons. (a) A representative 3D reconstruction of a rat hippocampal neuron expressing Lifeact-ruby (white) and the automated trace generated by Filament Tracer (blue). Scale bar is 10 μm. (b) Automated and manual spine counts were performed within the same dendritic regions. Top: A dendritic segment of a Lifeact-ruby-expressing neuron (white) and the corresponding automated trace (dendrite: red, spines: blue) as well as manually marked spines (yellow spheres) are shown. Middle: Lifeact-ruby signal (white) is overlayed by the automated trace (shown alone at right). Bottom: Lifeact-ruby signal (white) is overlayed by manual spine marks (yellow; shown alone at right). (c) For each dendritic segment, the manual spine count was plotted against the automated count. Linear regression analysis showed that manual and automated spine detection were significantly correlated (n = 28; b = 0.824; p < 0.001). Image and caption reprinted from Molecular Brain.
Their automated image analysis method produces accurate 3D reconstructions of spines without any manual validation, which can be used to analyze fixed or live neurons imaged with widefield fluorescence or confocal microscopy. The Bassell group used it to analyze changes in spine morphology after applying brain-derived neurotrophic factor (BDNF) to hippocampal neurons. BDNF is known to encourage growth and differentiation of new neurons and synapses, and the group's analysis revealed that BDNF induced rapid changes in spine neck and length geometry. They also showed that a potential therapeutic drug could rescue the established spine abnormalities of neurons isolated from a mouse model of fragile X syndrome, an inherited intellectual disability.
In the future, the researchers would like to make use of Imaris and Filament Tracer to investigate the fluorescence intensity of dendritic spine heads. “We would like to use this feature to assess trafficking of macromolecules into spines, and perhaps, correlate the trafficking of synaptic molecules with morphological parameters,” Dr. Bassell said. “In addition, we would like to continue using pharmacological treatments in disease models to better understand the underlying disease mechanisms and find potential drug therapies.”
Research Paper: Automated 4D analysis of dendritic spine morphology: applications to stimulus-induced spine remodeling and pharmacological rescue in a disease model, Molecular Brain. 2011, 4:38 doi:10.1186/1756-6606-4-38
