Hardware Solutions
Applications
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Cell Division
Zahi Abdul-Sater, Donna Cerabona and colleagues, Indiana University School of Medicine
Researchers including Zahi Abdul-Sater and Donna Cerabona, led by Grzegorz Nalepa, M.D., Ph.D., at Indiana University School of Medicine are studying the Fanconi anemia (FA/BRCA) pathway, a signaling network involving at least 21 proteins that maintain genomic stability, control growth and development, and prevent cancer. By combining super-resolution and deconvolution microscopy with Imaris software, the researchers have revealed further details of the biological mechanisms involved in this pathway’s ability to suppress tumors. The work could one day lead to the development of new strategies for fighting cancers involving this pathway.
Bi-allelic germline disruption of any FA gene causes Fanconi anemia, a genetic disorder characterized by developmental abnormalities, bone marrow failure, myelodysplasia, and increased cancer risk. Even in people without this disorder, heterozygous mutations in the FA genes increase risk of breast and ovarian cancers as well as other tumors.
Scientists know that the FA/BRCA pathway is important for DNA repair during the interphase stage of cell division. However, the Nalepa lab wanted to explore the role of this pathway beyond interphase. “We demonstrated that FANCA, an FA protein, is important for several mitotic processes like centrosome maintenance, microtubule nucleation, and spindle assembly checkpoint function,” said Abdul-Sater.
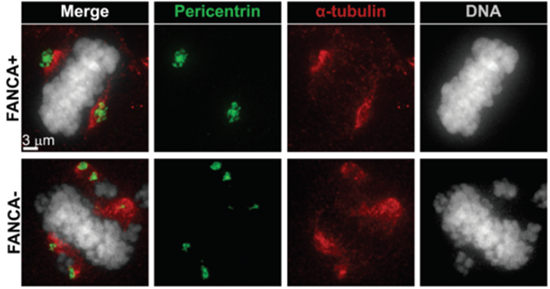
This figure shows increased centrosome number (marked in green) in a FANCA deficient patient mitotic cell (lower panel) compared to control (upper panel). The mitotic spindle is shown in red. The researchers opened deconvolved image stacks in Imaris 3D mode to visualize the total number of centrosomes in the cell. Centrosomes were then quantified and representative images were exported using Imaris Snapshot function. Courtesy of Zahi Abdul-Sater and Donna Cerabona, Indiana University School of Medicine.
Analyzing super-resolution images
The researchers said that Imaris is vital to work done in the Nalepa lab, which relies on super-resolution and deconvolution microscopy to study mitosis. They regularly use the software for image analysis and routinely employ the Imaris Spots 3D function to quantify recruitment of proteins to the kinetochore during mitosis.
“The intuitive user interface and the quantitative capabilities that Imaris provides are essential for our studies,” said Cerabona. “In this new study, Imaris was most useful in quantifying the number of microtubules emanating from the centrosome and the length of each individual microtubule.”
The Nalepa lab used the Imaris Snapshot mode to export the image stacks used in the published figures and used Imaris to assess mitotic spindle formation. To study spindle formation, the researchers acquired image stacks with a deconvolution microscope using 20x, 60x, or 100x lenses. The images were deconvolved and then opened in Imaris and viewed in “slice” mode. The researchers then scrolled through the slices to count the number of microtubules emanating from the centrosome and used the Line Measurement function to measure the length of individual spindles.
The Imaris image analysis showed that loss of FANCA impaired mitotic spindle assembly. These results combined with other analyses showed that FA/BRCA signaling serves as a type of gatekeeper by maintaining genomic integrity throughout interphase and mitosis. These new findings could lead to mitosis-targeted therapy for cancers involving the FA/BRCA network.
Research Paper: Abdul-Sater Z, Cerabona D, Potchanant ES, Sun Z, Enzor R, He Y, Robertson K, Goebel WS, Nalepa G. FANCA safeguards interphase and mitosis during hematopoiesis in vivo. Exp Hematol. 2015 Dec;43(12):1031-1046.e12
