Hardware Solutions
Applications
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Development
Mary E. Dickinson’s research group at the Baylor College of Medicine in Houston, Texas, studies morphogenesis during the early development of vertebrate embryos. The researchers developed a new method for mounting mouse embryos in agarose cylinders and demonstrated that it allows 24-hour time-lapse imaging with light-sheet microscopy. Using Imaris software, they tracked cell movement in these developing embryos.
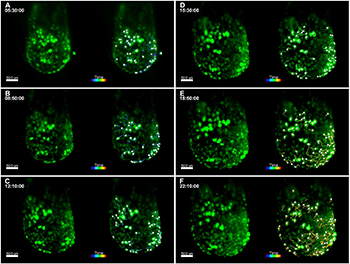
Live imaging of TCF/LEF-H2B::eGFP E6.5 embryos using agarose cylinders and 3D tracking of endothelial cells. For all panels, images from select time points from the 3D time-lapse movie are on the left and images from the movie overlaid with Imaris 3D tracking data are on the right. Embryos are oriented with the anterior side on the left. (A-F) Cultured embryos in hollow agarose cylinders show a twofold increase in embryo size over 24 h. Images from select time points are shown (A, 5 h 30 min; B, 8 h 50 min; C, 12 h 10 min; D, 15 h 30 min; E, 18 h 50 min; F, 22 h 10 min), and 3D cell tracks represent cell movements that occurred during the 2.5-h time period before the respective time point. Images from early time points show passive, radial cell movements associated with embryo growth (A-C), while images from later time points (E-F) show distinct antero-lateral migration events of deeper TCF/LEF-H2B::eGFP+ cells as gastrulation begins. GFP+ endoderm cells on the surface show less movement. Images were taken with a 20× objective, NA=1.0, zoom=0.7, every 10 min for a total of 24 h, z-step size=1.14 µm, total z-depth=115 µm. Scale bars: 50 µm.
Developmental biologists study morphogenesis to learn how the organism develops, specifically how the shape of organs and tissues form. Imaging embryo development provides scientists with quantitative data that can be used to prove how various morphogenesis events occur. Understanding these biological processes and how they go awry can provide insight into developmental disorders.
Imaging embryo development
Light-sheet microscopy is a popular technique for imaging embryo development. It uses a sheet of light to excite a thin plane of fluorescence in the sample, reducing out-of-focus fluorescence and improving image resolution. Three-dimensional images can be created by moving the sample so that the light sheet crosses the sample at different z-depths to form a z-stack or by rotating the embryo and acquiring images at various angles to create a 3D tomograph.
With light-sheet microscopy, the sample must be fixed in place in a way that it can be rotated for access from multiple sides. Hanging the specimen from a pin or embedding it in agarose can be problematic for delicate samples, such as post-implantation mouse embryos, that grow significantly and are sensitive to mounting. To solve these issues, the researchers developed hollow agarose cylinders that accommodate for embryonic growth while still providing boundaries that minimize tissue drift and enable imaging in multiple orientations.
Eliminating growth constraints
“The advantage to performing experiments using agarose cylinders is that it allows for embryos to expand in size as the embryo grows, and, therefore, eliminates constraints on growth imposed by embedding directly in the agarose,” said Ryan S. Udan, a postdoctoral fellow in the lab, who is now at Missouri State University. “The suspension of the embryo and ability to move the embryo in 3D, gives the added benefit of imaging the embryo at different angles with light sheet microscopy, including angles that are not easily accessible using a confocal microscopy system.”
To test the method, the researchers mounted mouse embryos in the agarose cylinders and conducted 24-h time-lapse imaging using light sheet microscopy. They acquired 66 to 200 z-slices for up to 100 time points, which created datasets ranging from 87 to 264 gigabytes in size. The images revealed many brightly fluorescent cells undergoing division and moving within the embryo. To analyze cell movement, they used the spot analysis function of Imaris software to analyze GFP expressing cells in 572 z-sections that represented images taken through half of the embryo from one angle. Due the large file sizes, the images were converted to one-channel, 8-bit TIF files, prior to cell tracking.
“Imaris can handle large amounts of data and allowed us to track the fluorescent nuclei in 3D,” Udan said. “Our experiments were a proof-of-principle that the approach of using agarose cylinders does work. Now that we have the ability to track cells in 3D, this can provide more accurate descriptions of embryo morphogenesis that have not been able to be addressed previously.”
Using this new approach, the researchers imaged various aspects of mouse embryo development, including dorsal aortae fusion and the expansion of the edge of the yolk sac as it expanded around the embryo, both of which can’t bee seen with confocal imaging because of difficultly in orienting the embryos. The researchers say that the hollow agarose cylinders can be used to image other types of embryos and tissue explants.
Author: Mary E. Dickinson, Ryan S. Udan, and colleagues, Baylor College of Medicine in Houston
Category: Case Study
